PUBLICATIONS
C. Paternoster, T. Tarenzi, R. Potestio, G. Latttanzi, Gamma-Hemolysin Components: Computational Strategies for LukF-Hlg2 Dimer Reconstruction on a Model Membrane, Int. J. Mol. Sci. (2023)

R. Fiorentini, T. Tarenzi and R. Potestio, Fast, accurate, and system-specific variable-resolution modelling of proteins, (2023)
2022 arXiv preprint here

M. Micheloni, L. Petrolli, G. Lattanzi R. Potestio, Kinetics of radiation-induced DNA double-strand breaks through coarse-grained simulations, Biophys. J (2023)
2022 biorXiv preprint here

R. Holtzman, M. Giulini, R. Potestio, Making sense of complex systems through resolution, relevance, and mapping entropy, Phys Rev E (2022)
2022 arXiv preprint here
M. Mele, R. Covino, R. Potestio, Information-theoretical measures identify accurate low-resolution representations of protein configurational space, Soft Matter (2022)
Featured in the themed collection Soft Matter Emerging Investigator Series
2022 arXiv preprint here
M. Rigoli, G. Spagnolli, G. Lorengo, P. Monti, R. Potestio, E. Biasini and A. Inga, Structural Basis of Mutation-Dependent p53 Tetramerization Deficiency, International Journal of Molecular Sciences (2022)
2022 biorXiv preprint here
T. Tarenzi, G. Mattiotti, M. Rigoli and R. Potestio, In search of a dynamical vocabulary: a pipeline to construct a basis of shared traits in large-scale motions of proteins, Applied Sciences (2022)
2022 biorXiv preprint here
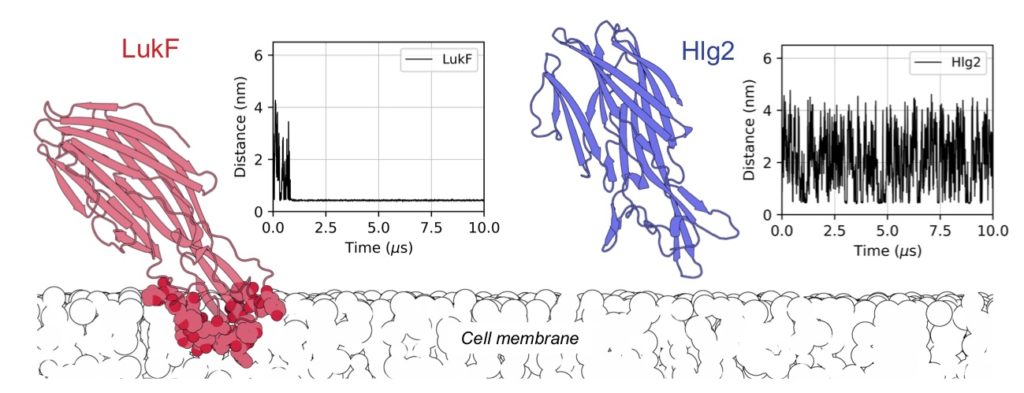
T. Tarenzi, G. Lattanzi and R. Potestio, Membrane binding of pore-forming γ-hemolysin components studied at different lipid compositions, BBA Biomembranes (2022)
2022 biorXiv preprint here
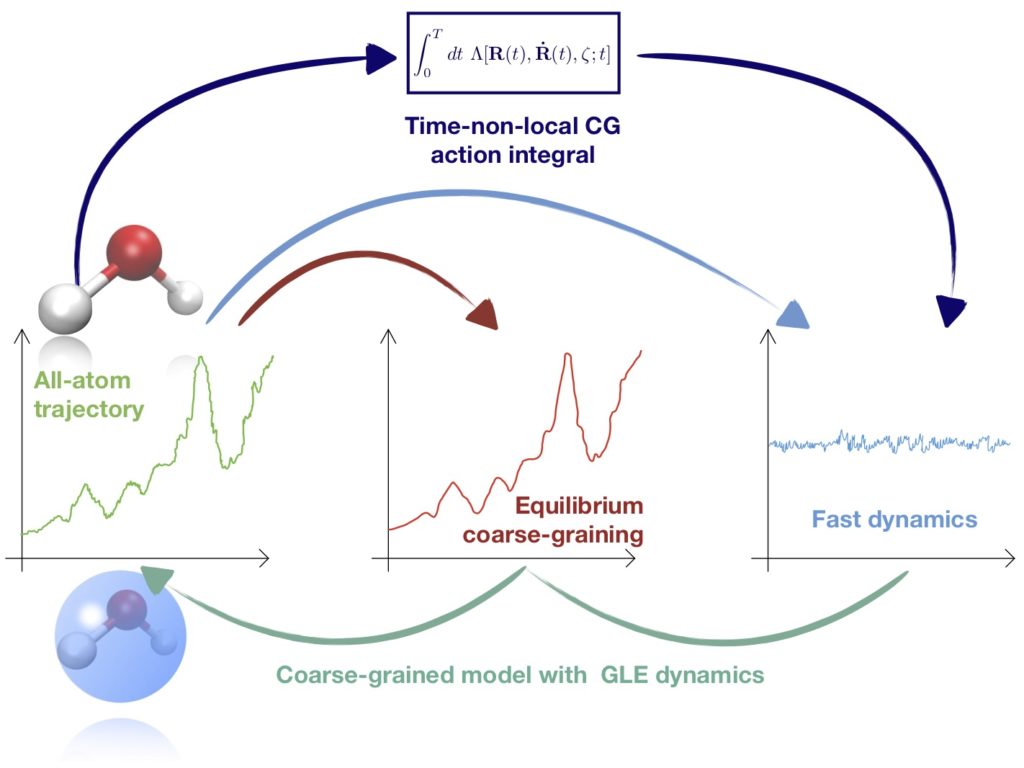
P. Luchi, R. Menichetti, G. Lattanzi and R. Potestio, Coarse-grained Mori-Zwanzig dynamics in a time-non-local stationary-action framework, arXiv preprint (2022)
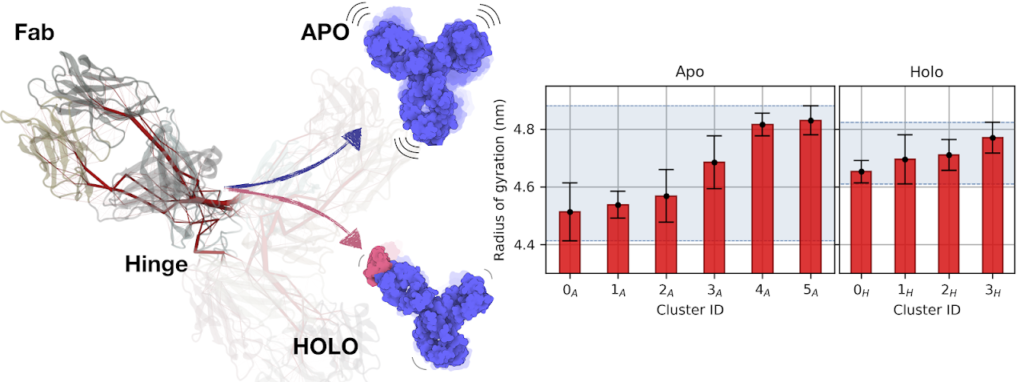
T. Tarenzi, M. Rigoli and R. Potestio, Communication pathways bridge local and global conformations in an IgG4 antibody, Scientific Reports (2021)
Link to the University of Trento press release (in Italian) here
2021 biorxiv preprint here
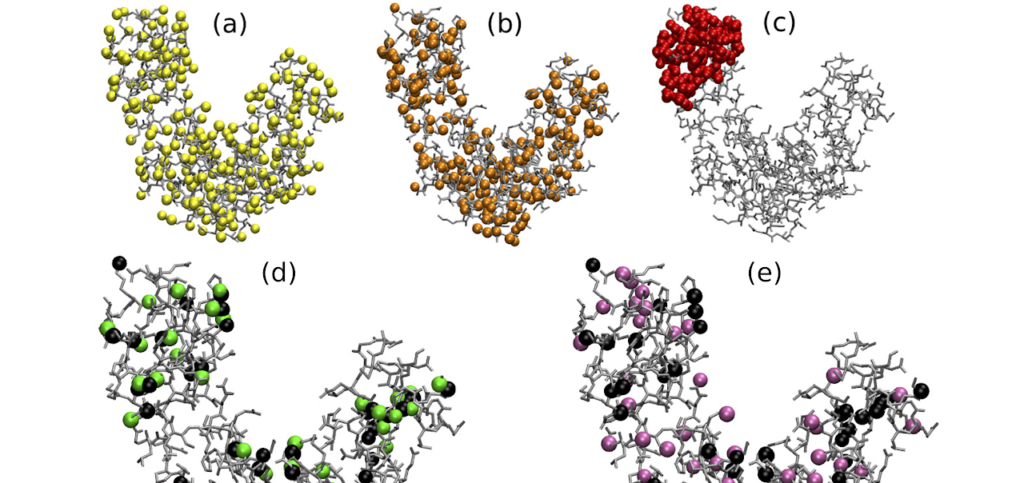
R. Menichetti, M. Giulini and R. Potestio, A journey through mapping space: characterising the statistical and metric properties of reduced representations of macromolecules, EPJB (2021)
2021 arxiv preprint here

M. Giulini, M. Rigoli, G. Matiotti, R. Menichetti, T. Tarenzi, R. Fiorentini and R. Potestio, From system modelling to system analysis: the impact of resolution level and resolution distribution in the computer-aided investigation of biomolecules, Front. Mol. Biosci. (2021)
F. Errica, M. Giulini, D. Bacciu, R. Menichetti, A. Micheli and R. Potestio, A deep graph network-enhanced sampling approach to efficiently explore the space of reduced representations of proteins, Front. Mol. Biosci. (2021)
2020 arxiv preprint here
L. A. Baptista, R. Chandra Dutta, M. Sevilla, M. Heidari, R. Potestio, K. Kremer and R. Cortes-Huerto, Density-Functional-Theory Approach to the Hamiltonian Adaptive Resolution Simulation Method, Journal of Physics: Condensed Matter (2021)
M. Giulini, R. Menichetti, M. Scott Shell and R. Potestio, An information theory-based approach for optimal model reduction of biomolecules, J. Chem. Theor. Comput. (2020) [See Resources section for open access raw data]
R. Fiorentini, K. Kremer and R. Potestio, Ligand-protein interactions in lysozyme investigated through a dual-resolution model, Proteins: structure, function, and bioinformatics (2020) [See Resources section for open access raw data]
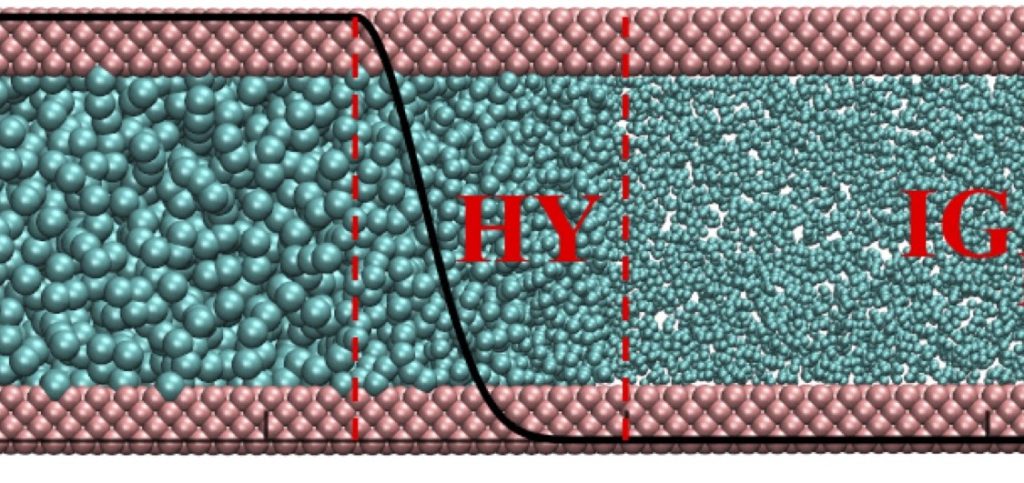
M. Heidari, K. Kremer, R. Golestanian, R. Potestio, and R. Cortes-Huerto, Open-Boundary Hamiltonian adaptive resolution. From grand canonical to non-equilibrium molecular dynamics simulations, J. Chem. Phys. (2020)

M. Giulini and R. Potestio, A deep learning approach to the structural analysis of proteins, Royal Society Interface Focus (2019) [See Resources section for open access raw data]

E. Riccardi, S. Pantano and R. Potestio, Envisioning data sharing for the biocomputing community, Royal Society Interface Focus (2019)
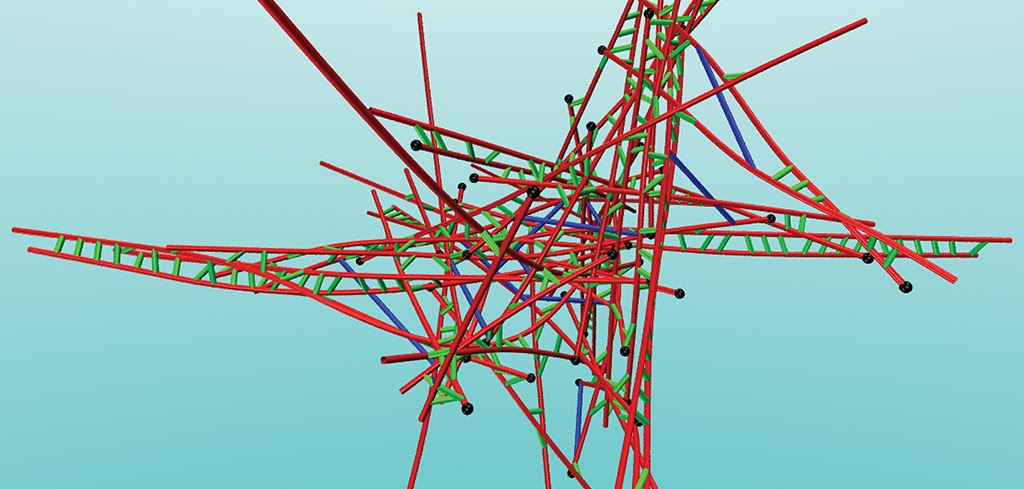
R. Erban, S. Harris and R. Potestio, Multi-resolution simulations of intracellular processes, Royal Society Interface Focus (2019)
Introduction to the Interface Focus theme issue Multi-resolution simulations of intracellular processes organised by Radek Erban, Sarah Harris and Raffaello Potestio
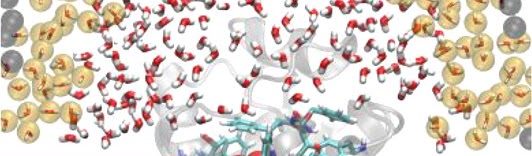
T. Tarenzi , V. Calandrini, R. Potestio, and P. Carloni, Open-Boundary Molecular Mechanics/Coarse-Grained Framework for Simulations of Low-Resolution G-Protein-Coupled Receptor–Ligand Complexes, J. Chem. Theor. Comput. (2019)
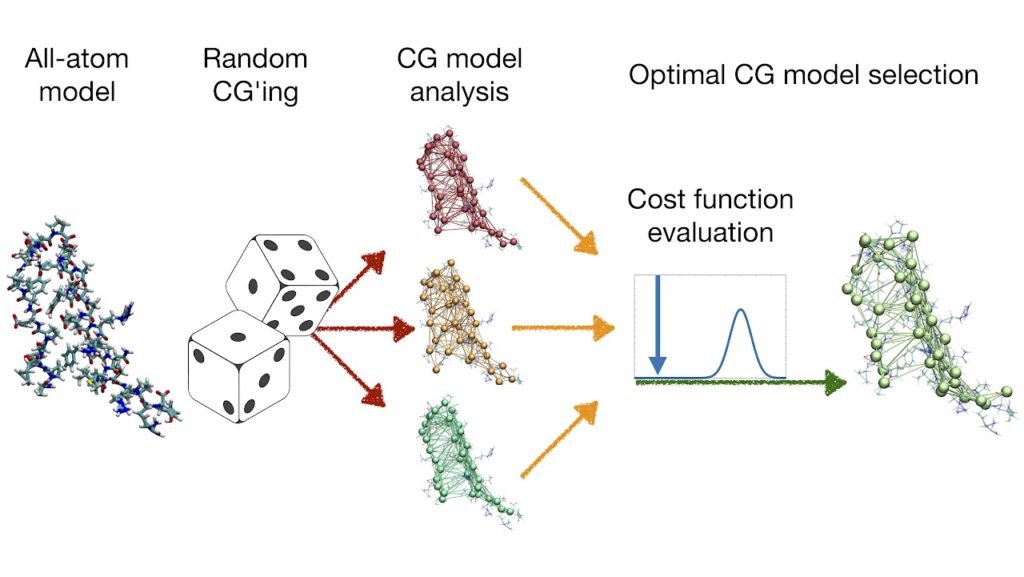
P. Diggins, C. Liu, M. Deserno, and R. Potestio, Optimal coarse-grained site selection in elastic network models of biomolecules, J. Chem. Theor. Comput. (2019)
RESOURCES

Raw data for the manuscript Membrane binding of pore-forming γ-hemolysin components studied at different lipid compositions, BBA Biomembranes (2022)
LINK to the Zenodo/OpenAIRE repository

Raw data for the manuscript Communication pathways bridge local and global conformations in an IgG4 antibody, Scientific Reports (2021)
LINK to the Zenodo/OpenAIRE repository

Raw data for the manuscript A mapping space Odyssey: characterising the statistical and metric properties of reduced representations of macromolecules, EPJB (2021)
LINK to the Zenodo/OpenAIRE repository

Raw data for the manuscript An information theory-based approach for optimal model reduction of biomolecules, J. Chem. Theor. Comput. (2020)
LINK to the Zenodo/OpenAIRE repository

Raw data for the manuscript Ligand-protein interactions in lysozyme investigated through a dual-resolution model, Proteins (2020)
LINK to the Zenodo / OpenAIRE repository

Raw data for the manuscript A deep learning approach to the structural analysis of proteins, Royal Society Interface Focus (2019)
LINK to the Zenodo / OpenAIRE repository
LINK to the GoogleDrive repository